Exercise 2b Filtering#
Aim: The purpose of this lab is to get you started thinking about frequency content of time series
Learning outcomes: At the end of this lab, you will be able to:
Filter data using windows
Check that the window used has “good” spectral properties (a good frequency response)
Data: You will work with a tide gauge time series.
Directions: Answer the questions, create an *.ipynb and three figures. Pick one of these figures to add to the shared_figures/ folder.
See also
Jonathan Lilly’s course on time series analysis for more about spectra: https://www.jmlilly.net/course/index.html.
Create a notebook & load the data#
Create an
*.ipynbcontaining the commands for this assignment, or copy this file.
Name your python notebook something useful ex<X>-<slug>-<Lastname>-seaocn.ipynb where you replace <X> with the exercise number and <slug> with the short slug to name the topic, and <Lastname> with your last name.
Figures should be named something like `ex<X>fig<Y>-<Lastname>-<slug>-seaocn.png` where you replace `<X>` with the exercise number, `<Y>` with the figure number, and `<Lastname>` with your last name.
```
Import necessary packages.
For example,
matplotlibandpandasandnumpyandxarray. You may also needIf you are missing any of these packages, please refer to Resources: Python.
# Importing required packages here
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
import pandas as pd
import datetime
# We are going to need some extra packages here
import scipy.signal as sg #Package for signal analysis
from scipy.fft import fft, fftshift
plt.rcParams["figure.figsize"] = (10,6.5) #set default figure size
plt.rcParams['font.size'] = 14
plt.rcParams['font.family'] = 'serif'
Download some data + quick check what’s there#
In the time series exercise, you did some quick and easy filtering by making annual averages. Now we’ll try something a little more complicated using tide gauge data.
Pick a tide gauge from the east coast of the USA: https://www.bodc.ac.uk/data/hosted_data_systems/sea_level/international/north_atlantic/#Duck and download the data in
*.ncformat.Put the file in your
data/directory.Load the data using
xarrayinto something calledtd.Squeeze the data in
xarrayto remove extra dimensions of your data variable.Print the data to see what is in there.
Make a simple plot of your data variable
sea_level_1against time.
# Update the code below for your data
file_path = '../data/'
filename = 'w9006.nc'
fname = file_path + filename
# Load the data
td = xr.open_dataset(fname)
td = td.squeeze()
#print(td)
# Quick plot data
#plt.plot(td.time,td.sea_level_1)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/file_manager.py:209, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
208 try:
--> 209 file = self._cache[self._key]
210 except KeyError:
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/lru_cache.py:55, in LRUCache.__getitem__(self, key)
54 with self._lock:
---> 55 value = self._cache[key]
56 self._cache.move_to_end(key)
KeyError: [<class 'netCDF4._netCDF4.Dataset'>, ('/Users/eddifying/Library/Mobile Documents/com~apple~CloudDocs/Work/teaching/seaocn-teaching/2024.seaocn-uhh/coursebook-seaocn-2024/coursebook_seaocn/seaocn-cb/data/w9006.nc',), 'r', (('clobber', True), ('diskless', False), ('format', 'NETCDF4'), ('persist', False)), '09542e29-f5d1-45e2-810a-d487ede7cfd9']
During handling of the above exception, another exception occurred:
FileNotFoundError Traceback (most recent call last)
Cell In[2], line 7
4 fname = file_path + filename
6 # Load the data
----> 7 td = xr.open_dataset(fname)
8 td = td.squeeze()
9 #print(td)
10
11 # Quick plot data
12 #plt.plot(td.time,td.sea_level_1)
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/api.py:541, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, backend_kwargs, **kwargs)
529 decoders = _resolve_decoders_kwargs(
530 decode_cf,
531 open_backend_dataset_parameters=backend.open_dataset_parameters,
(...)
537 decode_coords=decode_coords,
538 )
540 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 541 backend_ds = backend.open_dataset(
542 filename_or_obj,
543 drop_variables=drop_variables,
544 **decoders,
545 **kwargs,
546 )
547 ds = _dataset_from_backend_dataset(
548 backend_ds,
549 filename_or_obj,
(...)
557 **kwargs,
558 )
559 return ds
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/netCDF4_.py:578, in NetCDF4BackendEntrypoint.open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, mode, format, clobber, diskless, persist, lock, autoclose)
557 def open_dataset(
558 self,
559 filename_or_obj,
(...)
574 autoclose=False,
575 ):
577 filename_or_obj = _normalize_path(filename_or_obj)
--> 578 store = NetCDF4DataStore.open(
579 filename_or_obj,
580 mode=mode,
581 format=format,
582 group=group,
583 clobber=clobber,
584 diskless=diskless,
585 persist=persist,
586 lock=lock,
587 autoclose=autoclose,
588 )
590 store_entrypoint = StoreBackendEntrypoint()
591 with close_on_error(store):
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/netCDF4_.py:382, in NetCDF4DataStore.open(cls, filename, mode, format, group, clobber, diskless, persist, lock, lock_maker, autoclose)
376 kwargs = dict(
377 clobber=clobber, diskless=diskless, persist=persist, format=format
378 )
379 manager = CachingFileManager(
380 netCDF4.Dataset, filename, mode=mode, kwargs=kwargs
381 )
--> 382 return cls(manager, group=group, mode=mode, lock=lock, autoclose=autoclose)
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/netCDF4_.py:329, in NetCDF4DataStore.__init__(self, manager, group, mode, lock, autoclose)
327 self._group = group
328 self._mode = mode
--> 329 self.format = self.ds.data_model
330 self._filename = self.ds.filepath()
331 self.is_remote = is_remote_uri(self._filename)
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/netCDF4_.py:391, in NetCDF4DataStore.ds(self)
389 @property
390 def ds(self):
--> 391 return self._acquire()
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/netCDF4_.py:385, in NetCDF4DataStore._acquire(self, needs_lock)
384 def _acquire(self, needs_lock=True):
--> 385 with self._manager.acquire_context(needs_lock) as root:
386 ds = _nc4_require_group(root, self._group, self._mode)
387 return ds
File ~/micromamba/envs/seaocn_env/lib/python3.8/contextlib.py:113, in _GeneratorContextManager.__enter__(self)
111 del self.args, self.kwds, self.func
112 try:
--> 113 return next(self.gen)
114 except StopIteration:
115 raise RuntimeError("generator didn't yield") from None
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/file_manager.py:197, in CachingFileManager.acquire_context(self, needs_lock)
194 @contextlib.contextmanager
195 def acquire_context(self, needs_lock=True):
196 """Context manager for acquiring a file."""
--> 197 file, cached = self._acquire_with_cache_info(needs_lock)
198 try:
199 yield file
File ~/micromamba/envs/seaocn_env/lib/python3.8/site-packages/xarray/backends/file_manager.py:215, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
213 kwargs = kwargs.copy()
214 kwargs["mode"] = self._mode
--> 215 file = self._opener(*self._args, **kwargs)
216 if self._mode == "w":
217 # ensure file doesn't get overridden when opened again
218 self._mode = "a"
File src/netCDF4/_netCDF4.pyx:2464, in netCDF4._netCDF4.Dataset.__init__()
File src/netCDF4/_netCDF4.pyx:2027, in netCDF4._netCDF4._ensure_nc_success()
FileNotFoundError: [Errno 2] No such file or directory: '/Users/eddifying/Library/Mobile Documents/com~apple~CloudDocs/Work/teaching/seaocn-teaching/2024.seaocn-uhh/coursebook-seaocn-2024/coursebook_seaocn/seaocn-cb/data/w9006.nc'
Filter the data#
Select a one year time period to work with#
Pick a year that looks filled in. I.e., straight lines between big blocks of color are probably because there’s not data.
Take a look at the attributes to try to verify that your year has no gaps
Subselect the data using the
.loccommand. For example, if you would like to extract the month of January in 1995, you would do:
# Your code here
Fig. 1 Calculate a moving average (Quick-and-dirty filtering)#
You will use xarray.DataArray.rolling()
From your one year period, plot a two week period. As an example, the code below plots data from a 1-year extract, and then adjusts the x-axis limits to be January 1-14, 1995.
Calculate a rolling average (moving average) over 5 hours from your dataset, and add this to your plot in red. Note that your data are assumed to be hourly.
Recalculate this using the option
center=True, and add this to your plot in green.Plot your figure and name it fig1 using the file naming convention.
What is the difference between using center=False or True?
# Your code here
Experiment with windows#
A better filter choice is a ‘windowed’ moving average. It smoothes the data around the edges of the window by weighting the edges less and the middle of the window higher.
Run the code below and take a look at the different windows (weighting) that can be used. Here we are using a window length of 100.
Create a window. When we are using a default rolling average, it is a rectangular or “boxcar” window (where “boxcar” refers to a train car that looks like a container on a container ship). Note that when importing packages above, we imported
scipy.signalassg.Personal favourite: Tukey. When you use it on a dataset, it looks a lot like a boxcar, but less jaggedy, and the spectral properties (frequency response) are good. You can take a quick look at the “frequency response” for different windows by clicking the function in the
scipydocumentation. If the side lobes drop away quickly and smoothly (to low decibels) from the center peak, this is better.
See also
Window types in scipy: https://docs.scipy.org/doc/scipy/reference/signal.windows.html#module-scipy.signal.windows
# Set a window length for this demo code
NN = 100
# Create a few window types.
myboxcar = sg.boxcar(NN)
myhanning = sg.hann(NN)
mytukey = sg.tukey(NN)
myparzen = sg.parzen(NN)
fig, axs = plt.subplots(2)
axs[0].plot(myboxcar)
axs[0].plot(myhanning)
axs[0].plot(mytukey)
axs[0].plot(myparzen)
wins = [myboxcar, myhanning, mytukey, myparzen]
for window in wins:
A = fft(window, 2048) / (len(window)/2.0)
freq = np.linspace(-0.5, 0.5, len(A))
response = 20 * np.log10(np.abs(fftshift(A / abs(A).max())))
axs[1].plot(freq, response)
axs[1].axis([-0.5, 0.5, -120, 0])
/var/folders/t1/z5bp59k95119nw35yqv699t40000gn/T/ipykernel_28737/1134310159.py:21: RuntimeWarning: divide by zero encountered in log10
response = 20 * np.log10(np.abs(fftshift(A / abs(A).max())))
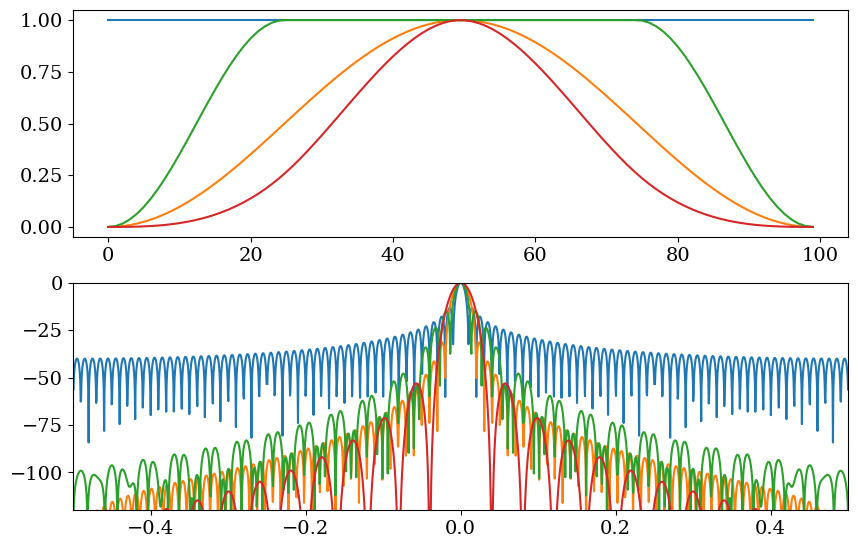
Fig. 2 Calculate moving averages with different filters#
This is a tide gauge record, so let’s think about removing the tides. Most tides that we normally think about are diurnal or semi-diurnal, so we’ll start by smoothing the data with a 1-day filter.
Recalculate your moving averages using two types of filters. One of these should be a boxcar, and the other is your choice.
Plot the two filtered time series and the original data on a new set of axes, and zoom in to a 2-week period for better visibility.
Export this figure as fig2.
However! This won’t work on xarray since their rolling doesn’t appear to support windows. So we will need to convert the data to a pandas dataframe first.
```{python}
sla_df = td_1995.sea_level_1.to_dataframe()
type(sla_df)
print(sla_df)
```
Then see https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.rolling.html for how to use the optional argument win_type.
Pandas rolling input doesn’t take time=24 but instead just the number 24 as the specified window length. Since our dataset is hourly, then a window of length 24 is a 24-hour window. If our data were monthly, how long would the filter be?
After plotting your time series, note which is bumpier (has higher standard deviation), the one with the boxcar filter or your other window? Think about why this might be.
# Your code here
Fitting a harmonic (optional)#
More typically, to remove tides, the tidal constituents can be removed using a harmonic fit, followed by e.g. a 40-hour filter. For a boxcar, this would be a 40-hour boxcar. For a tukey, it would be something longer.
Explore: See whether you can fit a harmonic to your tidal time series. What are the major tidal constituents?
Some packages exist to help with this: e.g., moflaher/ttide_py
Explore: Go a little deeper into time series analysis by considering spectra. Spectra can immediately help you identify dominant frequencies in your time series, whether they are seasonal (a 365-day cycle) or tidal (12- or 12.4-hr). An example notebook can be found here: https://www.jmlilly.net/course/labs/html/SpectralAnalysis-Python.html from Jonathan Lilly’s time series analysis course.
# Your code here (optional)
Fig. 3 Regridding data#
Now suppose you need to compare your tidal time series with a daily temperature record (for example). You want to calculate a linear regression, but the lengths of the time grids are different. You have some options.
Average your data into 1 day averages. Try this using the methods from the previous exercise.
Subsample your data into the value nearest to noon each day.
Linearly interpolate your data onto the time dimension of the temperature record.
Try these methods and consider what the differences might mean.
For the purpose of this exercise, simply create these new time series and plot a week or a month’s worth of data with all three time series on the same plot.
Save this figure as fig3 using the file naming convention.
# Your code here
